March 16, 2004
Thesauri, SKOS and terminology variation
There's a lot of activity in Semantic-Web-Land these days. I've been skeptical about the prospects for this work (e.g. here and here), but I'll be happy for any success that these folks manage to achieve, and I try to stay current. Here's a note on some SW doings, which you may find interesting if you're in the same sort of boat that I am.
SKOS-core 1.0 is "an RDF schema for representing thesauri and similar types of knowledge organisation system (KOS)", being developed by SWAD-Europe. Here's the current version of the SKOS-core 1.0 rdf file. Its "sister vocabulary" SKOS-mapping "allows you to assert mappings between concepts from different schemes". This is a picture of the "meta-model" of SKOS, showing two schemata of concepts with a partial mapping between them and labels for some of the concepts:
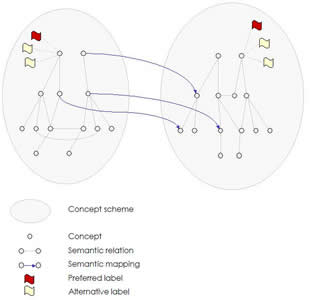
The SKOS-Core Guide says that
SKOS-Core is intended as a complement to OWL. It does provide a basic framework for building concept schemes, but it does not carry the strictly defined semantics of OWL. Thus it is ideal for representing those types of KOS, such as thesauri, that connot be mapped directly to an OWL ontology. SKOS is also easier to use, and harder to misuse than OWL, providing an ideal entry point for those wishing to use the Semantic Web for knowledge organisation.
Here's a recent W3C press release about OWL, in case you're not up on current Semantic Web acronyms.
My first point of reference for this stuff is a practical one -- how can I use it in projects that I'm involved with? For biomedical information extraction, terminology and terminology variation is a big issue, and so is connection of referents across different ontologies and ontology-like databases. So the issues that SKOS is addressing are relevant ones for some of the work that I do.
But so far, I'm not convinced that the SWAD-Europe work -- or any of the Semantic Web work -- is engaging these problems in a helpful or realistic way. The only example of terminological variation that I can find so far on their pages is
<rdf:RDF
xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:skos="http://www.w3.org/2004/02/skos/core#"
<skos:Concept rdf:about="urn:swad-e:example/concept/0001">
<skos:prefLabel>Bangers and mash</skos:prefLabel>
<skos:altLabel>Sausage and mash</skos:altLabel>
<skos:altLabel>Sausage and mashed potato</skos:altLabel>
<skos:inScheme rdf:resource="urn:swad-e:example/thesaurus"/>
</Concept></rdf:RDF>
Of course this is not intended to be anything more than a toy example to show how the system works, but it helps make clear that what SKOS is offering: the ability to designate a string as a preferred label for a concept, and a set of strings as alternative labels. However, real-world terminology variation usually looks like something other than just a list of alternative strings. Real-word terms are often complex phrases with free variation among alternatives in several different locations, with variable phrasing and ordering, drawn from a large and apparently open-ended set. Experience suggests that it's hard to get adequate coverage just with a set of strings, even with a quite large set of strings. It's not entirely clear what the best long-term approach will be, but a plausible way to get reasonable performance is to apply a statistical pattern recognition algorithm, trained on a set of examples in context and perhaps provided with a more general model of terminological variation. I'll give a simple example below in support of this view.
Note that this leaves aside several more difficult questions: the relationships among referents vs. the structure of the ontology, the problems of metonymy and synecdoche, elliptical variants of terms, etc. I'm talking about the easy case where there is a single well-defined referent and a bunch of strings that are clear and complete references to it. We can find some (relatively easy) examples of this kind by scanning the MEDLINE corpus for examples of explicitly defined acronyms. A typical source sentence containing three explicitly-defined acronyms:
Luteinizing hormone/chorionic gonadotropin (LH/CG) receptor complementary DNA (cDNA) isoforms were amplified using pseudopregnant rat ovarian total RNA as a template and the primers reaching over the coding regions at both ends in a reverse transcriptase-polymerase chain reaction (RT-PCR).
These should be cases where terminology is "on its best behavior", so to speak.
It's easy to recognize these patterns and map the acronyms onto the corresponding strings. When we look at the resulting sets of defining strings for a given acronym, they turn out to be remarkably diverse. The top of the histogram for definitions of RT-PCR is given below, with each example preceded by the count of occurrences (in our slightly-out-of-date local copy of MEDLINE). I haven't folded case or eliminated hyphens, but with or without such normalization, there are a lot of variants. This is just the head of the list -- there several times as many more to come, though with lower counts -- and one suspects that there are other variants in principle "out there", that didn't happen to come up in MEDLINE's billion words.
2191 reverse transcription-polymerase chain reaction
1627 reverse transcriptase-polymerase chain reaction
731 reverse transcription polymerase chain reaction
683 reverse transcriptase polymerase chain reaction
273 Reverse transcription-polymerase chain reaction
216 reverse-transcriptase polymerase chain reaction
211 reverse-transcription polymerase chain reaction
178 Reverse transcriptase-polymerase chain reaction
159 reverse transcription-PCR
123 reverse transcription and polymerase chain reaction
84 Reverse transcriptase polymerase chain reaction
80 Reverse transcription polymerase chain reaction
76 reverse transcriptase PCR
56 reverse transcription PCR
56 reverse transcriptase-PCR
26 Reverse transcription-PCR
25 reverse transcription followed by polymerase chain reaction
24 Reverse-transcription polymerase chain reaction
18 reverse-transcription-polymerase chain reaction
18 reverse transcription and the polymerase chain reaction
17 Reverse-transcriptase polymerase chain reaction
15 reverse-transcriptase-polymerase chain reaction
15 reverse-transcribed polymerase chain reaction
15 reverse transcribed polymerase chain reaction
13 Reverse transcriptase-PCR
11 Reverse transcriptase PCR
11 reverse transcribed-polymerase chain reaction
10 reverse transcription coupled to polymerase chain reaction
9 reverse transcription-polymerase chain reactions
9 Reverse Transcription-Polymerase Chain Reaction
9 Reverse transcription PCR
9 reverse-transcription PCR
9 Reverse transcription and polymerase chain reaction
9 reverse-transcriptase PCR
9 reverse transcriptase-linked polymerase chain reaction
8 reverse transcriptional polymerase chain reaction
8 reverse transcriptase-polymerase chain reaction
Let me make it clear that these particular instances are not problematic, since a constant acronym RT-PCR is given adjacent to them. We're interested in the problem of how to recognize instances of the "same" term in general, and this list just represents a convenient way to get a lot of examples of alternate complete renditions of a well-defined term in a well-controlled context. I'm not trying to insist that no finite list could possibly cover such cases adequately. However, a complete enough list would be quite long and quite hard to compile -- and probably the easiest way to compile it would be to use a generative model for terminological variation, effectively equivalent to the pattern-recognition approach that I've suggested as an alternative.
There's nothing wrong with providing a standard XML method for giving a thesaurus list of alternative strings for an item in an ontology. However, I think it's naive to suppose that this will be go very far towards solving the problem of recognizing "entity mentions" in texts and connecting them to standard referents, even in the simplest and most straightforward cases such as the one described above.
It's fair to respond that the authors of SKOS are trying to solve a different problem, namely how to let people who are putting explicit semantics in their web documents do so in a way that allows for variable concept labels and partly-related alternative conceptual schemata. Fine -- but some people may think that this will help to represent the content of the ordinary-language documents that ordinary folk write, especially when the documents are scientific or technical in character. But it won't.
Posted by Mark Liberman at March 16, 2004 12:23 AM