October 10, 2006
Fable 2.0
Here's something new from one of my day jobs. FABLE ("Fast Automated Biomedical Literature Extraction") version 2.0 is now online at http://fable.chop.edu. The FABLE blurb:
FABLE performs document retrieval and gene list extractions of MEDLINE abstracts for queries of genes, transcripts, and proteins by annotating text in a completely automated process. The system is currently optimized for human genes. FABLE combines a named entity recognition extractor and a human gene normalizer that have been applied to all MEDLINE records. A query interface allows users to search for articles mentioning particular genes or proteins of interest, or to generate lists of genes mentioned in articles associated with any keyword(s). Identified articles and gene lists can be sorted in various ways, depending upon users' preference. FABLE also allows users to download articles and gene sets using a variety of formats.
FABLE was developed by the BioIE group at the University of Pennsylvania and the Children's Hospital of Philadelphia. FABLE and BioIE are supported in part through grants from the National Science Foundation and the National Institutes of Health.
The great new thing is a "Gene Lister" application that creates lists of relevant human genes from the biomedical literature, based on arbitrary boolean combination of keyword searches. Keywords can reference drugs, diseases, people, places, institutions, genes, proteins -- or any other text in the indexed literature.
Pete White's email announcing the application gives these as "for instance" searches:
Identify genes associated with a disease: {schizophrenia AND bipolar NOT depression}
Identify genes associated with a disease attribute: {metastasis AND "colon cancer"}
Identify genes associated with a person: {asthma AND "Doe JA"}
Identify genes associated with a place: {"University of Pennsylvania" AND "heart disease"}
Identify genes associated with genes: {myoglobin NOT hemoglobin}
Identify genes associated with techniques: {luciferase}
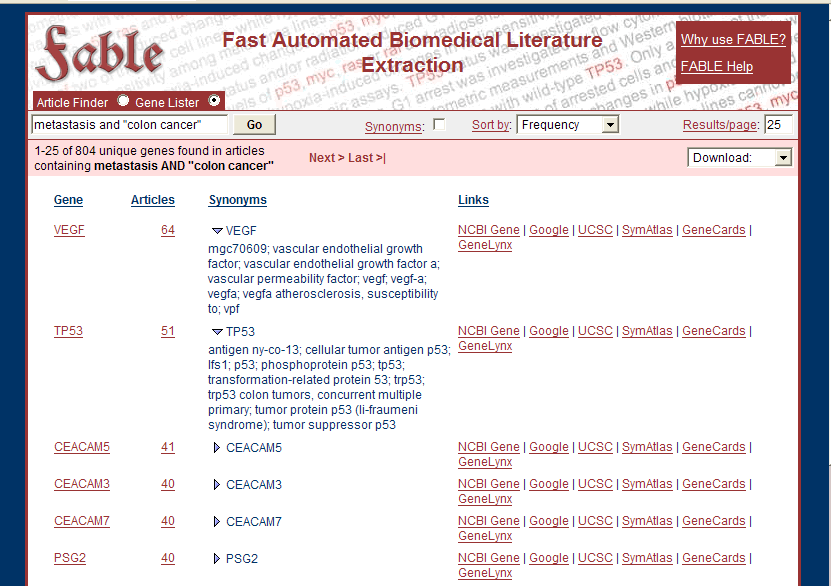
For example, if you type {metastasis AND "colon cancer"} into the "Gene Lister" field, you'll get something like this:
Note that you can click next to the gene symbols in the results, to see a list of synonyms.
The motivation for creating "Gene Lister" came from some experiments done last year by our collaborators at Children's Hospital. Here's a description of one of those experiments, from our yearly report to the National Science Foundation last summer:
We created a list of genes implicated in a specific biological process by applying our gene tagger and a rudimentary normalization process (case-insensitive exact string matching) to a set of 41,000 MEDLINE abstracts mentioning angiogenesis. A list of 2,460 genes extracted from and normalized in these documents was then compared against a manual list of 247 genes that was compiled by angiogenesis experts.
All but 2 of the genes in the manual list were also identified by the text-mined list. The text-mined list was relevance ranked based on the number of documents in which each gene was mentioned, and the 247 highest-ranking genes were compared with the comparably-sized manual list for precision. All of the 50 highest-ranked text-mined genes were identified as being legitimately associated with angiogenesis after further literature review. Furthermore, article recall was 17-fold higher than for articles linked to genes in Entrez Gene, and gene recall was 95-fold higher than for genes assigned to angiogenesis-related GO terms through AMIGO, demonstrating the current under-annotation of these resources for human genes.
The comparably-sized manual and text-mined lists were then compared by their respective correlations with gene expression profiles in low stage (rarely angiogenic) and advanced stage (angiogenic) neuroblastomas; by their correlations with protein pathways preferentially implicated in cancer; and with Gene Ontology annotations. In each case, the text-mined gene list correlated more closely with angiogenesis than did the manual list. Furthermore, blind evaluation of the highest-ranked text-mined genes by biomedical domain experts determined that the text-mined predictions were more accurate than expert opinion.
Importantly, the results of this exercise were deemed successful by the domain expert, as a slightly edited version of the text-mined list is now being used by Dr. X's lab as an initial screen for genes of interest in neuroblastoma progression. These results indicate that even a completely unsupervised process of compiling gene lists performs at high accuracy with our system.
The technology in the "Gene Lister" application is improved in several ways over what was used in the experiment described -- the gene tagger is somewhat better and the normalizer is a lot better -- so we have high hopes that it will be useful.
The data indexed by FABLE is refreshed weekly from MEDLINE®/PubMed®.
Posted by Mark Liberman at October 10, 2006 10:15 PM