October 19, 2006
The grammar of anti-microbial peptides
Several people have written to ask me about the work recently in the news under headlines like "Grammar helps biologists invent 40 bacteria fighters", "Grammatical rules spell out new drugs", "Grammar may help fight bacteria", "Bacteria, meet your new enemy: Grammar", "Scientists use grammar rules to slay killer germs", and (my personal favorite) "DaVinci Code meets CSI in bacteria battle".
So let's go directly to the paper itself: Christopher Loose, Kyle Jensen, Isidore Rigoutsos and Gregory Stephanopoulos, "A linguistic model for the rational design of antimicrobial peptides", Nature 443, 867-869(19 October 2006). Here's the abstract:
Antimicrobial peptides (AmPs) are small proteins that are used by the innate immune system to combat bacterial infection in multicellular eukaryotes1. There is mounting evidence that these peptides are less susceptible to bacterial resistance than traditional antibiotics and could form the basis for a new class of therapeutic agents. Here we report the rational design of new AmPs that show limited homology to naturally occurring proteins but have strong bacteriostatic activity against several species of bacteria, including Staphylococcus aureus and Bacillus anthracis. These peptides were designed using a linguistic model of natural AmPs: we treated the amino-acid sequences of natural AmPs as a formal language and built a set of regular grammars to describe this language. We used this set of grammars to create new, unnatural AmP sequences. Our peptides conform to the formal syntax of natural antimicrobial peptides but populate a previously unexplored region of protein sequence space.
Before we get to the details, some crucial background is in order, which I'll provide in the form of some structured links to the wikipedia.
A protein is a "large organic compound made of amino acids arranged in a linear chain and joined by peptide bonds". There are 20 "standard amino acids" used in proteins by living things on earth, and for convenience, these can be put into correspondence with the 20 letters {ARNDCEQGHILKMFPSTWYV}. Thus the primary structure of any protein can be specified as a single string of letters drawn from this alphabet. (Most proteins then fold up in complicated secondary, tertiary and quaternary structures, and these folded structures are crucial to their function, but that's another story).
Formal language theory, as developed by linguists, mathematicians and computer scientists during the second half of the 20th century, deals with the properties of (typically infinite) sets of (typically finite) strings made up of elements drawn from a (typically finite) vocabulary. Formal grammars are used to specify the properties of particular formal languages. Such grammars, and the languages they specify, can be arranged in a hierarchy of increasingly powerful types, known as the Chomsky hierarchy (because it was first worked out by Noam Chomsky and Marcel-Paul Schützenberger). The simplest, most limited, easiest-to-process type of grammar in this hierarchy is a regular grammar (also sometimes called a finite-state grammar).
Because proteins (and other biological macromolecules such as DNA and RNA) are finite-length strings of elements drawn from a finite alphabet, it's natural to describe them grammatically. And as a result, computational biology has long since borrowed the mathematical and computational tools of computational linguistics, and made good use of them in applications from gene-finding to protein-structure prediction. One of the first researchers to see the potential of such methods was David B. Searls, one of the founders of the Center for Bioinformatics at Penn, and now at GSK; a good source of insight is his 2002 Nature review article "The language of genes" (or try this link).
Continuing with the Loose et al. paper:
Our preliminary studies of natural AmPs indicated that their amphipathic structure gives rise to a modularity among the different AmP amino-acid sequences. The repeated usage of sequence modules—which might be a relic of evolutionary divergence and radiation—is reminiscent of phrases in a natural language, such as English. For example, the pattern QxEAGxLxKxxK (where 'x' is any amino acid) is found in more than 90% of the insect AmPs known as cecropins. On the basis of this observation, we modelled the AmP sequences as a formal language—a set of sentences using words from a fixed vocabulary. In this case, the vocabulary is the set of naturally occurring amino acids, represented by their one-letter symbols.
We conjectured that the 'language of AmPs' could be described by a set of regular grammars. [...]
Specifically, they decided to explore the hypothesis that AmPs are texts made up of 10-letter sentences:
To find a set of regular grammars to describe AmPs we used the Teiresias pattern discovery tool. With Teiresias, we derived a set of 684 regular grammars that occur commonly in 526 well-characterized eukaryotic AmP sequences from the Antimicrobial Peptide Database (APD). Together, these ~ 700 grammars describe the 'language' of the AmP sequences. In this linguistic metaphor, the peptide sequences are analogous to sentences and the individual amino acids are analogous to the words in a sentence. Each grammar describes a common arrangement of amino acids, similar to popular phrases in English. For example, the frog AmP brevinin-1E contains the amino-acid sequence fragment PKIFCKITRK, which matches the grammar P[KAYS][ILN][FGI]C[KPSA][IV][TS][RKC][KR] from our database (the bracketed expression [KAYS] indicates that, at the second position in the grammar, lysine, alanine, tyrosine or serine is equally acceptable). On the basis of this match, we would say that the brevinin-1E fragment is 'grammatical'.
By design, each grammar in this set of ~ 700 grammars is ten amino-acids long and is specific to AmPs—at least 80% of the matches for each grammar in Swiss-Prot/TrEMBL (the APD is a subset of Swiss-Prot/TrEMBL) are found in peptides annotated as AmPs.
They then used a simple method to find new "grammatical" sequences to synthesize:
To design unnatural AmPs, we combinatorially enumerated all grammatical sequences of length twenty (see Methods) in which each window of size ten in the 20-mers was matched by one of the ~ 700 grammars. We chose this length because we could easily chemically synthesize 20-mers and this length is close to the median length of AmPs in the APD. From this set, we removed any 20-mers that had six or more amino acids in a row in common with a naturally occurring AmP; we then clustered the remaining sequences on the basis of similarity, choosing 42 to synthesize and assay for antimicrobial activity.
They created some obvious control sequences:
For each of the 42 synthetic peptides, we also designed a shuffled sequence, in which the order of the amino acids was rearranged randomly so that the sequence did not match any grammars. These shuffled peptides had the same amino-acid composition as their synthetic counterparts and thus, the same molecular weight, charge and isoelectric point (bulk physiochemical factors that are often correlated with antimicrobial activity). We hypothesized that because the shuffled sequences were 'ungrammatical' they would have no antimicrobial activity, despite having the same bulk physiochemical characteristics. In addition, we selected eight peptides from the APD as positive controls and six 20-mers from non-antimicrobial proteins as negative controls.
Then they tested their 42 synthetic peptides, along with the controls, against Bacillus cereus and Escherichia coli, "as representative gram-positive and gram-negative bacteria". Here's their schematic diagram of the method:
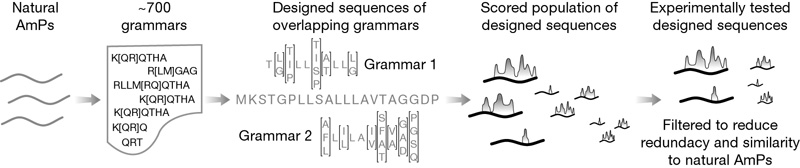
And here's their summary of the results:
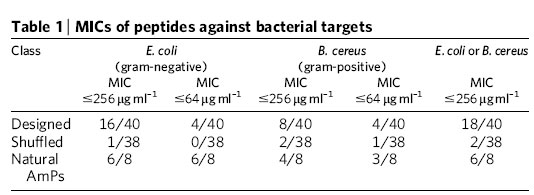
In general, the reports in the popular press are not too bad, though I don't think that most of them really give readers much of a sense of what was really done and why it made sense to try it. As usual, the AP story scrapes the bottom of the science-journalism barrel exhibits extraordinary interpretive creativity:
Biologists reached back to elementary school to discover a promising new way to fight nasty bacteria: Apply the rules of grammar.
The unusual method to try to defeat drug-resistant microbes and anthrax borrows a page from ``The Da Vinci Code'' and the TV show ``CSI: Crime Scene Investigation.''
But the kind of grammar that kids (used to) learn in elementary school has almost nothing in common but a name with the kind of grammar that Loose et al. used. And it never would have occurred to me that you could help readers to understand the role of formal language theory in this piece of research by reference to those two estimable works of popular culture, The Da Vinci Code and CSI. But now that I come to think of it, I can see that this method of getting science across to the public has real legs:
Physicists reached out to the National Football League to find a way to create the heaviest element yet.
Or:
To understand the recent Hawaiian earthquakes, seismologists borrowed a page from "American Idol" and "Pirates of the Caribbean".
What does it mean? Who knows. Does it sell papers? Apparently.
[By the way, not everyone is impressed by the Loose et al. paper. For example, Derek Lowe at In the Pipeline writes that
There's nothing here that any drug company's bioinformatics people wouldn't be able to do for you, as far as I can see.
So why haven't they? Well, despite the article's mention of a potential 50,000 further peptides of this type, the reason is probably because not many people care. After all, we're talking about small peptides here, of the sort that are typically just awful candidates for real-world drugs.
I'm not going to try to evaluate either the novelty or the value of the work. But I do think that it's interesting to try to figure out why this particular letter to Nature got such a comparatively big play in the world's media. Is "grammar" really all that catchy a hook?]
[Update -- Fernando Pereira writes
I feel that the use of the term "grammar" in the paper is misleading. Sure, these regular patterns are grammars in the formal sense, but they yield finite languages, so there isn't any anything there different from the standard techniques (eg. position-dependent weight matrices) for representing such motifs in computational biology. Their use of Teiresias is pretty standard too for that particular tool. Teiresias can only discover non-recurring (Kleene star-free) patterns yielding finite languages. This is quite adequate for motif discovery, but would be useless for cases with recurring (eg. some transmenbrane proteins) or recursive (eg.certain RNA and protein folds) structure. Those were the applications of formal grammar that were discussed in David Searls' pioneering paper, not mere motif discovery.
That's a fair criticism. Of course, it applies to the paper's authors and to the editors of Nature, not to the popular press, who would have been just as happy to make an unenlightening connection to the kind of grammar that Americans used to learn in "grammar school", even if the paper in question had been about patterns that required grammars rather than position-dependent weight matrices to describe.]
Posted by Mark Liberman at October 19, 2006 11:30 AM