September 21, 2007
Wherein I take the bait
Coturnix at Blog Around the Clock has the interesting job of "Online Community Manager" at PLoS-ONE (Public Library of Science). "My job", he tells us, "is to try to motivate you to comment on the papers there". Doing his job yesterday with respect to a recently-published PLoS paper by Gang Li et al., "Accelerated FoxP2 Evolution in Echolocating Bats", he wrote
Looking forward to further responses by other blogs, hopefully Afarensis, John Hawks and Language Log?
I don't have much time this morning, so I'll start by pointing people to our earlier posts on the subject, and echo the suggestion that people read the excellent and quite accessible new PLoS paper -- which everyone can do, because PLoS is Open Access! Here's the new paper's abstract:
FOXP2 is a transcription factor implicated in the development and neural control of orofacial coordination, particularly with respect to vocalisation. Observations that orthologues show almost no variation across vertebrates yet differ by two amino acids between humans and chimpanzees have led to speculation that recent evolutionary changes might relate to the emergence of language. Echolocating bats face especially challenging sensorimotor demands, using vocal signals for orientation and often for prey capture. To determine whether mutations in the FoxP2 gene could be associated with echolocation, we sequenced FoxP2 from echolocating and non-echolocating bats as well as a range of other mammal species. We found that contrary to previous reports, FoxP2 is not highly conserved across all nonhuman mammals but is extremely diverse in echolocating bats. We detected divergent selection (a change in selective pressure) at FoxP2 between bats with contrasting sonar systems, suggesting the intriguing possibility of a role for FoxP2 in the evolution and development of echolocation. We speculate that observed accelerated evolution of FoxP2 in bats supports a previously proposed function in sensorimotor coordination.
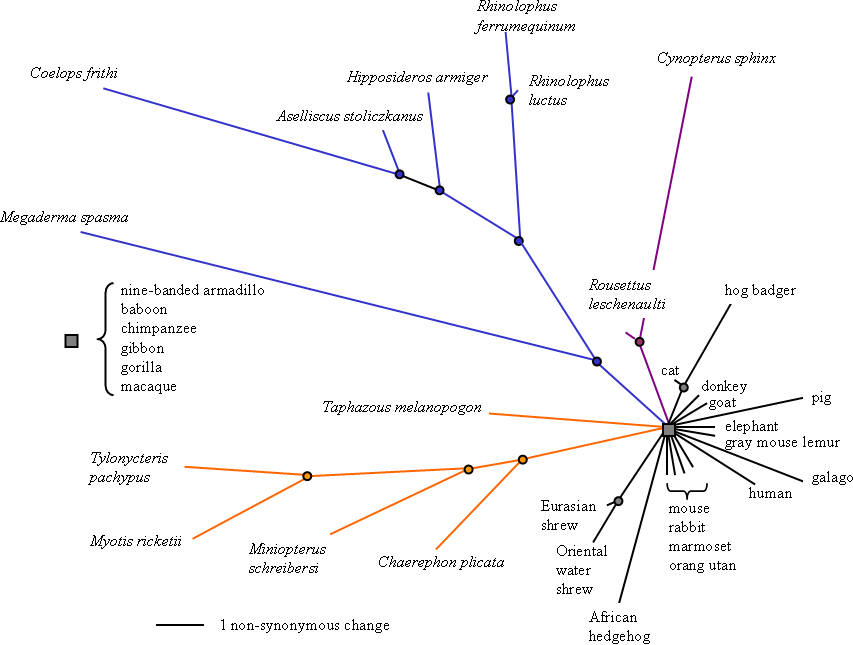
I'll also reproduce their cool phylogenetic tree (actually it's more of a phylogenetic thicket, or perhaps a phylogenetic tumbleweed) showing that whatever use humans have made of our little FOXP2 innovation, it's small change relative to the massive innovations by the bats in the same gene:
Figure 1. Radial phylogenetic tree showing relative rates of non-synonymous evolution among 35 eutherian mammals, including 13 bats.
Bats species are given as italicised binomial names. Branch lengths based on maximum-likelihood estimates of non-synonymous substitutions along 1995 bp of the FoxP2 gene are superimposed onto a cladogram based on published trees. Bat lineages are coloured to show the echolocating Yinpterochiroptera (blue) that mostly possess high duty constant frequency (CF) calls with at least partial Doppler shift compensation, the Yangochiroptera (orange) that mostly possess low duty cycle calls, as well as the absence of laryngeal echolocation in Yinpterochiroptera fruit bats (violet).
And I'll also put in another plug for FABLE ("Fast Automated Biomedical Literature Extraction"), which is the result of an NSF-funded collaboration between computational linguists at Penn and biomedical researchers at Children's Hospital. It's fast, it's free, and it'll let you find all the original research article that mention FOXP2 under any of its synonyms (cag repeat protein 44; cagh44; dkfzp686h1726; forkhead box p2; forkhead box protein p2; foxp2; spch1; tnrc10; trinucleotide repeat-containing gene 10; trinucleotide repeat-containing gene 10 protein), or search for other genes often mentioned in association with FOXP2 (it finds 99 of them, starting with PRM2, FOXP1, FOXP3, IL2, SHC2, AUTS1), and so on.
And finally, I'd like to head off a mis-interpretation. Gang Li et al. write that FOXP2 is "implicated in the development and neural control of orofacial coordination, particularly with respect to vocalisation". They don't intend the implication that FOXP2 should be identified as (something like) "the gene for orofacial sensorimotor coordination" (even though that would be a step up from calling it "the language gene" or "the grammar gene" -- see Christine Kenneally, "First Language Gene Found", Wired News, 2001). They don't intend this implication because they know very well than many other genes are also "implicated in the development and neural control of orofacial coordination",a nd that FOXP2 is a transcription factor that affects the expression of many other genes (or better, is part of many other genetic networks) with many other functions as well.
Thus Weiguo Shu et al., "Foxp2 and Foxp1 cooperatively regulate lung and esophagus development", Development 134: 1991-2000, 2007:
The airways of the lung develop through a reiterative process of branching morphogenesis that gives rise to the intricate and extensive surface area required for postnatal respiration. The forkhead transcription factors Foxp2 and Foxp1 are expressed in multiple foregut-derived tissues including the lung and intestine. In this report, we show that loss of Foxp2 in mouse leads to defective postnatal lung alveolarization, contributing to postnatal lethality. Using in vitro and in vivo assays, we show that T1alpha, a lung alveolar epithelial type 1 cell-restricted gene crucial for lung development and function, is a direct target of Foxp2 and Foxp1. [...] These data identify Foxp2 and Foxp1 as crucial regulators of lung and esophageal development, underscoring the necessity of these transcription factors in the development of anterior foregut-derived tissues and demonstrating functional cooperativity between members of the Foxp1/2/4 family in tissues where they are co-expressed.
There's obviously some interesting connection between FOXP2 and vocalization, as you can learn by reading Stephanie A. White et al., "Singing Mice, Songbirds, and More: Models for FOXP2 Function and Dysfunction in Human Speech and Language", The Journal of Neuroscience, 26(41):10376-10379, 2006. But let's not get carried away.
Post-finally, I'll end by expressing some puzzlement about what seems to be a careless mis-statement in the Blog Around The Clock post that I'm responding to. It starts this way:
Earlier studies have indicated that a gene called FOXP2, possibly involved in brain development, is extremely conserved in vertebrates, except for two notable mutations in humans. This finding suggested that this gene may in some way be involved in the evolution of language, and was thus dubbed by the popular press "the language gene". See, for instance, this and this for some recent research on the geographic variation of this gene (and related genes) and its relation to types of languages humans use (e.g., tonal vs. non-tonal).
The "geographical variation" business refers to the work by Dediu and Ladd that was previously discussed on Language Log here and here. They correlated the population frequency of adaptive haplogroups of the two genes, ASPM and Microcephalin, with the geographical distribution of lexical tone. (Well, actually, they correlated the geographic distribution of 1,000 genetic variants with the geographic distribution of 26 linguistic traits, but that was only to provide a background distribution for evaluating the correlation of Microcephalin and ASPM with lexical tone.)
Now, neither Microcephalin nor ASPM is another name for FOXP2. They're not even close. The UCSC Gene Browser (accessed via FABLE's LitTrack feature) identifies ASPM as chr1:195,319,996-195,386,439 (i.e. positions 195,319,996 to 195,386,439 on chromosome 1); Microcephalin as MCPH1 at chr8:6,251,528-6,509,561; and FOXP2 as chr7:113,513,617-114,157,642.
What was Coturnix thinking? I'd hate to think that he was just stirring the pot with random evocative references, as less reputable PR operations are wont to do.
[Update -- Bora Zivkovic, aka Coturnix, wrote (in part):
Thank you for the excellent response. I have to admit that I put my post together in great haste (thus my mistake in it, but I was hoping to start a discussion) [...]
Translation: He was stirring the pot with random evocative references -- but it was in a good cause ;-)! ]
Posted by Mark Liberman at September 21, 2007 06:16 AM